Point process neuron with double-exponential shaped PSCs. More...
#include <poisson_dbl_exp_neuron.h>
Public Member Functions | |
| PoissonDblExpNeuron () | |
| PoissonDblExpNeuron (const PoissonDblExpNeuron &) | |
| nest::port | send_test_event (nest::Node &, nest::rport, nest::synindex, bool) |
| void | handle (nest::SpikeEvent &) |
| void | handle (nest::CurrentEvent &) |
| void | handle (nest::DataLoggingRequest &) |
| nest::port | handles_test_event (nest::SpikeEvent &, nest::rport) |
| nest::port | handles_test_event (nest::CurrentEvent &, nest::rport) |
| nest::port | handles_test_event (nest::DataLoggingRequest &, nest::rport) |
| void | get_status (DictionaryDatum &) const |
| void | set_status (const DictionaryDatum &) |
 Public Member Functions inherited from spore::TracingNode Public Member Functions inherited from spore::TracingNode | |
| TracingNode () | |
| virtual | ~TracingNode () |
| void | get_trace_status (DictionaryDatum &d) const |
| const_iterator | get_trace (nest::delay steps, trace_id id) const |
| Access the trace of id at time step step. More... | |
| const_iterator | get_trace (nest::Time const &time, trace_id id) const |
| size_t | get_num_traces () const |
Friends | |
| class | nest::RecordablesMap< PoissonDblExpNeuron > |
| class | nest::UniversalDataLogger< PoissonDblExpNeuron > |
Additional Inherited Members | |
 Public Types inherited from spore::TracingNode Public Types inherited from spore::TracingNode | |
| typedef CircularBuffer< double >::const_iterator | const_iterator |
| typedef size_t | trace_id |
 Protected Member Functions inherited from spore::TracingNode Protected Member Functions inherited from spore::TracingNode | |
| void | init_traces (size_t num_traces) |
| void | set_trace (nest::delay steps, double v, trace_id id=0) |
Point process neuron with double-exponential shaped PSCs.
PoissonDblExpNeuron is a variant of the spike response model model with double exponential PSP shapes of the from 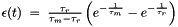 and absolute refractory mechanism.
and absolute refractory mechanism.
Spikes are generated randomly according to the current value of the transfer function which operates on the membrane potential. Spike generation is followed by an optional dead time (refractory state). Setting with_reset to true will reset the membrane potential after each spike.
The transfer function can be chosen to be linear, exponential or a sum of both by adjusting three parameters:
![\[ rate = Rect[ c1 * V' + c2 * exp(c3 * V') ], \]](form_0.png)
where the effective potential 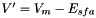 and
and 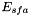 is called the adaptive threshold. By setting c3 = 0, c2 can be used as an offset spike rate for an otherwise linear rate model.
is called the adaptive threshold. By setting c3 = 0, c2 can be used as an offset spike rate for an otherwise linear rate model.
The dead time enables to include refractoriness. If dead time is 0, the number of spikes in one time step might exceed one and is drawn from the Poisson distribution accordingly. Otherwise, the probability for a spike is given by 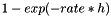 , where h is the simulation time step. If dead_time is smaller than the simulation resolution (time step), it is internally set to the time step. Note that, even if non-refractory neurons are to be modeled, a small value of dead_time, like dead_time=1e-8, might be the value of choice since it uses faster uniform random numbers than dead_time=0, which draws Poisson numbers. Only for very large spike rates (> 1 spike/h) this will cause errors.
, where h is the simulation time step. If dead_time is smaller than the simulation resolution (time step), it is internally set to the time step. Note that, even if non-refractory neurons are to be modeled, a small value of dead_time, like dead_time=1e-8, might be the value of choice since it uses faster uniform random numbers than dead_time=0, which draws Poisson numbers. Only for very large spike rates (> 1 spike/h) this will cause errors.
The model implements an adaptive threshold. If the neuron spikes, the threshold increases and the membrane potential will take longer to reach it. If the neuron does not spike, the threshold linearly decays over time, increasing the firing probability of the neuron (see [6], parameters: target_rate, target_adaptation_speed).
This model has been adapted from iaf_psc_delta. The default parameters are set to the mean values in [2], which have were matched to spike-train recordings.
Parameters
The following parameters can be set in the status dictionary. (default values and constraints are given in parentheses):
| name | type | comment |
|---|---|---|
| V_m | double | Membrane potential [mV] |
| tau_rise_exc | double | Fall time constant of excitatory PSP (2.0, >0.0) [ms] |
| tau_fall_exc | double | Rise time constant of excitatory PSP (20.0, >0.0) [ms] |
| tau_rise_inh | double | Fall time constant of inhibitory PSP (1.0, >0.0) [ms] |
| tau_fall_inh | double | Rise time constant of inhibitory PSP (10.0, >0.0) [ms] |
| dead_time | double | Duration of the dead time (1.0, &ge 0.0) [ms] |
| dead_time_random | bool | Should a random dead time be drawn after each spike? (false) |
| dead_time_shape | int | Shape parameter of dead time gamma distribution (1, &ge 1) |
| t_ref_remaining | double | Remaining dead time at simulation start (>0.0) ms |
| with_reset | bool | Should the membrane potential be reset after a spike? |
| I_e | double | Constant input current (0.0) [pA] |
| input_conductance | double | Conductance of input currents (1.0) [S] |
| c_1 | double | Slope of linear part of transfer function (0.0) [Hz/mV] |
| c_2 | double | Prefactor of exponential part of transfer function (1.238) [Hz/mV] |
| c_3 | double | Coefficient of exponential non-linearity of transfer function (0.25, &ge 0.0) [1/mV] |
| target_rate | double | Target rate of neuron for adaptation mechanism (10.0, &ge 0.0) [Hz] |
| target_adaptation_speed | double | Speed of rate adaptation (0.0, &ge 0.0) |
Sends: SpikeEvent
Receives: SpikeEvent, CurrentEvent, DataLoggingRequest
References
[1] Multiplicatively interacting point processes and applications to neural modeling (2010) Stefano Cardanobile and Stefan Rotter, Journal of Computational Neuroscience.
[2] Predicting spike timing of neocortical pyramidal neurons by simple threshold models (2006) Jolivet R, Rauch A, Luescher H-R, Gerstner W. J Comput Neurosci 21:35-49.
[3] Pozzorini C, Naud R, Mensi S, Gerstner W (2013) Temporal whitening by power-law adaptation in neocortical neurons. Nat Neurosci 16: 942-948. (uses a similar model of multi-timescale adaptation).
[4] Grytskyy D, Tetzlaff T, Diesmann M and Helias M (2013) A unified view on weakly correlated recurrent networks. Front. Comput. Neurosci. 7:131.
[5] Deger M, Schwalger T, Naud R, Gerstner W (2014) Fluctuations and information filtering in coupled populations of spiking neurons with adaptation. Physical Review E 90:6, 062704.
[6] David Kappel, Robert Legenstein, Stefan Habenschuss, Michael Hsieh and Wolfgang Maass. Reward-based self-configuration of neural circuits. 2017. https://arxiv.org/abs/1704.04238
| spore::PoissonDblExpNeuron::PoissonDblExpNeuron | ( | ) |
Default Constructor.
| spore::PoissonDblExpNeuron::PoissonDblExpNeuron | ( | const PoissonDblExpNeuron & | n | ) |
Copy Constructor.
|
inlinevirtual |
Status getter function.
Reimplemented from spore::TracingNode.
| void spore::PoissonDblExpNeuron::handle | ( | nest::SpikeEvent & | e | ) |
SpikeEvent handling.
| e | the event. |
| void spore::PoissonDblExpNeuron::handle | ( | nest::CurrentEvent & | e | ) |
CurrentEvent handling.
| e | the event. |
| void spore::PoissonDblExpNeuron::handle | ( | nest::DataLoggingRequest & | e | ) |
DataLoggingRequest handling.
| e | the event. |
|
inline |
PoissonDblExpNeuron test event.
|
inline |
PoissonDblExpNeuron test event.
|
inline |
PoissonDblExpNeuron test event.
|
inline |
PoissonDblExpNeuron test event.
|
inlinevirtual |
Status setter function.
Reimplemented from spore::TracingNode.
 1.8.13
1.8.13